
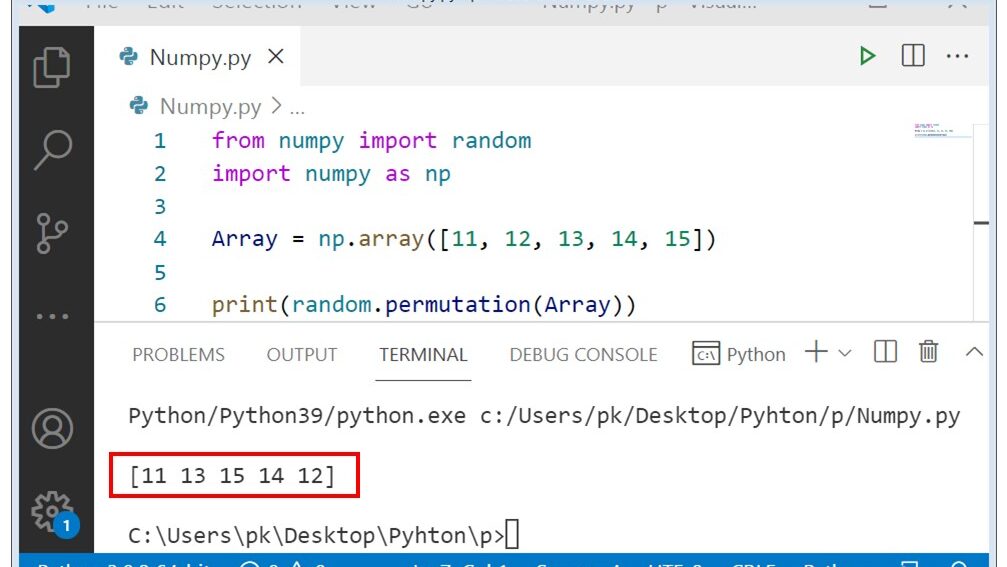
- #MATLAB RANDOM PERMUTE MATRIX HOW TO#
- #MATLAB RANDOM PERMUTE MATRIX DOWNLOAD#
- #MATLAB RANDOM PERMUTE MATRIX MAC#
If omitted, all observations are treated as exchangeable and belonging to a single large exchangeability block. Default is 10000.Įxchangeability blocks file, in csv or vest format. Use -n 0 to run all permutations and/or sign-flips exhaustively. Run only the F-contrasts, not the t-contrasts. The option -f cannot be used more than the number of times the option -t is used. Each file supplied with a -f corresponds to the file supplied with the option -t immediately before. The option -f can be used more than once, so that more than one F-contrasts file can be loaded. The option -t can be used more than once, so that more than one t-contrasts file can be loaded.į-contrasts file, in csv or vest format. T-contrasts file, in csv or vest format (the format used by FSL).
#MATLAB RANDOM PERMUTE MATRIX HOW TO#
For information on how to construct the design matrix, see the FSL GLM manual.

It can be in csv format, or in fsl's vest format. Alternatively, if the areas are not meaningful for cluster extent or TFCE, this argument can be simply a number, such as "1", which is then used as the area of all vertices or faces.ĭesign matrix. If the second argument is given, it should contain the areas, which are then used (e.g., average areas from native geometry after areal interpolation). If only the surface file is provided, its area is calculated and used for the computation of spatial statistics (cluster extent and TFCE). The second is an optional area-per-vertex or area-per-face file, or simply a number. The first argument is the surface file itself. This option is needed when the input data is a scalar field over a surface and cluster extent or TFCE have been enabled. When more than one is supplied, each -s should be entered in the same order as the respective -i. Either one for all inputs, or one per input supplied in the same order as the respective -i appear. Except for NPC and MV, mixing is allowed (e.g., voxelwise, vertexwise and non-imaging data can be all loaded at once, and later will be all corrected across). All input files must contain the same number of observations (e.g., the same number of subjects).
More than one can be specified, each one preceded by its own -i. This can be done with the command addpath:
Uncompress the downloaded file, start Octave, and add the newly created directory to the Octave path. Typing palm at the prompt without arguments shows usage information. Uncompress the downloaded file, open Matlab, and add the newly created directory to the Matlab path (menu File -> Set Path). If you are using Mac, and choose Octave, note that reading of NIFTI files need the option '-noniiclass' to work properly (more details below). The path can also be added to the system's $PATH variable, so that it can be easily called from any directory just by typing palm. palm in the directory where it was installed. If the one that is chosen isn't in the $PATH variable, make sure to specify also the path to the directory that contains the executable for either of these (whichever you choose). This is a script, inside which you can set whether the script should use Octave or Matlab. Open the file palm (not to be confused with palm.m). It may be much simpler to run PALM as a command directly from the shell in Linux or in Mac, and it can easily be called from scripts. In Windows, it can be executed inside Matlab or Octave.
#MATLAB RANDOM PERMUTE MATRIX MAC#
In Linux and in Mac systems, it can be executed in any of these ways. PALM can run as a standalone command (i.e., executed directly from the command line/terminal), or inside Octave or Matlab.
#MATLAB RANDOM PERMUTE MATRIX DOWNLOAD#
To download latest packaged version, please click here.Īlternatively, visit the repository on GitHub.


 0 kommentar(er)
0 kommentar(er)
